T-DNA Primer Design
( Powered by GEBD )
|
Please use the backup page served by AtTA, if the tdnaexpress server is down.
The new T-DNA Primer Design Tool is now powered by Genome
Express Browser Server (GEBD). The new tool can return the primers faster, and also give the insertion
location information, the estimated T-DNA confirmation product size, as well as primer3-like format output.
(July 28, 2005) | |
|
| |
|
1. Protocol for SALK T-DNA primer design
|
 |
Note:
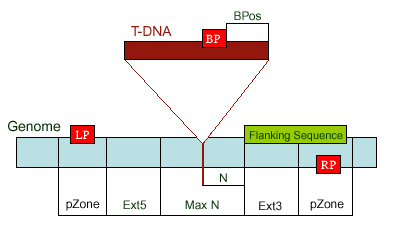
MaxN - Maximum difference of the actual insertion site and the sequence, default 300 bps pZone - Regions used to pick up primers, default 100 bps Ext5, Ext3 - Regions between the MaxN to pZone, reserved not for picking up primers LP, RP - Left, Right genomic primer BP - T-DNA border primer LB - the left T-DNA border primer BPos - The distance from BP to the insertion site LB - Left border primer of the T-DNA insertion:
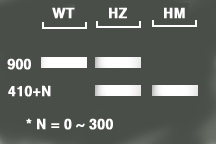
GCGTGGACCGCTTGCTGCAACT >LBb1.3 (Newly used by Salk Genotyping Project and with better results) ATTTTGCCGATTTCGGAAC >LBa1 of pBIN-pROK2 for SALK lines TGGTTCACGTAGTGGGCCATCG >LB_6313R for SALK lines TCAAACAGGATTTTCGCCTGCT >LB1 for SAIL lines C/418-451 of pCSA110-pDAP101_T-DNAs GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC > >LB2 for SAIL lines C/390-423 of pCSA110-pDAP101_T-DNAs GCTTCCTATTATATCTTCCCAAATTACCAATACA >LB3 for SAIL lines C/350-383 of pCSA110-pDAP101_T-DNAs TAGCATCTGAATTTCATAACCAATCTCGATACAC To download SAIL pCSA110 & pDAP101 T-DNAs. By using the three primers (LBb1.3+LP+RP) for SALK lines, users for WT (Wild Type - no insertion) should get a product of about 900-1100 bps ( from LP to RP ), for HM (Homozygous lines - insertions in both chromosomes) will get a band of 410+N bps ( from RP to insertion site 300+N bases, plus 110 bases from LBb1.3 to the left border of the vector), and for HZ (Heterozygous lines - one of the pair chromosomes with insertion) will get both bands. The product size should be 200 base larger if using LBa1 instead of LBb1.3. However, the protocol requires the same or similiar TM values for all the LB, LP and RP primers. You can set up two paired reactions, LP+RP and LB+RP. You should get a product in the LP+RP reaction for WT or HZ lines or get blank for HM lines, while get a band in the LB+RP for HM or HZ lines. |
|
2. SALK T-DNA verification primer design
The program will pick up LP and RP for you. Its product size is around 900-1100 bps. If you want to
confirm whether the design is right, simply cut and paste
the LP and RP sequences together (you can cut/paste whole returned primer info line) to the blast textarea on the tdnaexpress page, hit "Return" if only one line of text and submit it.
If you want to design primers within your sequences, please click here.
Now you could paste any T-DNA lines on the T-DNA Express to design their primers (LP/RP). However, 1) please use their own (left) border primer (LB) in PCR. 2) please re-set the parameter Ext5 or MAX_N value according to the distance from their LB to the insertion site. The new Ext5 or MAX_N = 300 + thedistance - 110. |
|
visits since Jan. 30, 2003 | |
| © SIGnAL 2000- | | Home | About Us | Microarray | cDNA | T-DNA | Database | Links | Contact Us | |