Arabidopsis sequence indexed TDNA insertion
-- Project FAQ --
Frequently Asked Questions
A User's Guide to the Arabidopsis T-DNA Insertional Mutant Collections
1. Q: Which
A:
2. Q: Which T-DNA border flanking sequences were
amplified and sequenced? What are the primers do you use for left border PCR?
A:
For PCR 1, we use LBa1 primer: 5' tggttcacgtagtgggccatcg 3'
For PCR 2 and sequencing, we use LBb1 primer: 5' gcgtggaccgcttgctgcaact 3'
3. Q: Is there any way to design a Right Border primer for PCR at the other end of the insertion? Is the right border less predictable in its insertion pattern?
A: A: A: A: A: T A: A: A: A: A: The arrow indicates the orientation of
the T-DNA insert within the chromosome. The arrow point in the 5' to 3'
direction beginning with the left border DNA and into the plant flanking
DNA sequence). A: The sequence of the
left border of pROK2 and the position of the PCR amplification and sequencing
primers: A: A:
4. Q: What is the number of T-DNA inserts per
line?
5. Q: Are multiple T-DNA insertions amplified
and detectable by sequencing?
6. Q: How can I obtain seeds for the Salk insertion
lines?
7. Q: The ABRC or NASC has sent me seeds for a Salk insertion
mutant. What generation (posttransformation) are these seeds?
8. Q: What is the plant selectable marker used
for the transformation?
9. Q: What is the T-DNA transformation vector
used to generate the mutant?
10. Q: Which T-DNA border is used for plant
flanking sequence isolation?
11. Q: Given that each Salk line
contains an average of 1.5 -2.0 TDNA insertions. How many sequenced products
be generated per line?
12. Q: On the T-DNA Express website
what are the arrows above the T-DNA line?
13. Q: What is the DNA sequence around
the left border of pROK2 and where do the PCR isolation and DNA sequencing
primers anneal?
6042 5'CTGATGGGCTGCCTGTAT
LBb1 primer
6061 CGAGTGGTGATTTTGTGCCGAGCTGCCGGTCGGGGAGCTGTTGGCTGGCTGGTGGCAGGA
6121 TATATTGTGGTGTAAACAAATTGACGCTTAGACAACTTAATAACACATTGCGGACGTTTT
6181
TAATGTACTGGGGTGGTTTTTCTTTTCACCAGTGAGACGGGCAACAGCTGATTGCCCTTC
6241
ACCGCCTGGCCCTGAGAGAGTTGCAGCAAGCGGTCCACGCTGGTTTGCCCCAGCAGGCGA
3'<<<<<<<<<<<<<<<<<<<<<<
5'
6301
AAATCCTGTTTGATGGTGGTTCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGC
6361
CCGAGATAGGGTTGAGTGTTGTTCCAGTTTGGAACAAGAGTCCACTATTAAAGAACGTGG
6421
ACTCCAACGTCAAAGGGCGAAAAACCGTCTATCAGGGCGATGGCCCACTACGTGAACCAT
3'<<<<<<<<<<<<<<<<<<<<<<
5'
LBa1 primer6481
CACCCAAATCAAGTTTTTTGGGGTCGAGGTGCCGTAAAGCACTAAATCGGAACCCTAAAG
6541 GGAGCCCCCGATTTAGAGCTTGACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGGA
6601 AGAAAGCGAAAGGAGCGGGCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTG
6661 CGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGT
6721 TGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGAG
6781 CTCGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGCGTAATCATG-3'
14. Q: What
is the sequence profile of the T-DNA inserts in the SIGnAL T-DNAExpress
database (and which has submitted to GenBank- GSS division)
5' T-DNA left-border
sequence¨¤ plant
genomic sequence¨¤
Linker/adaptor 3'
5' CTCTCGGGCCGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGG
TGAAAAGAAAAACCACCCCAGTACATTAAAAACGTCCGCAATGTGTTAT TAAGTTGTCTAAGCGTCAATTTGTTTACACCACAATATATCCT
15. Q: Is
sequencing of the entire T-DNA collection finished?
16. Q:
A:
17. Q: What are the items labeled-
RAFL_xxxx, Ceres_xxxx, AF_xxxx or AY_xxxx -within the TDNAExpress database?
A:
18. Q: I searched the TDNAExpress database
using a protein/DNA sequence of interest to my lab and got the following
result (below). What is the name of the Salk insertion line and how can
I order it?
CODE At1g01330
COD2 F6F3.31
CHRO chr1
TITL hypothetical protein
POSN 00125932
COOR C/125932-126589,126686-126840,126935-127453,127532-127573,127651-127782, 127868-127935,128028-128312,128479-128551,128968-129060,129853-129914, 130039-130099
HITS SALK_013537.54.95.x
CHRO chr1
POSN 127748
DIRE W/127748-127778
LOCA Exon
A:
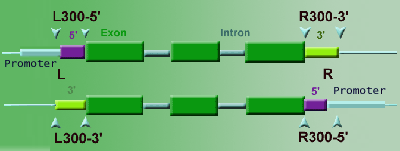
19. Q: Hi! I would like to ask you a couple of things, and for doing so I took one example from the T-DNA Express page:
gene At1g01060
Hit SALK_061415.55.50.x
LOCA L300-3'
1) 55.50 is the avg of the 20 best continuous base quality scores. Is
this a good quality value? how would you compare this value with a value,
let's say, of 13.80?
A: We use a program, Phred to read DNA sequencer trace data. It calls bases and assigns quality values to the bases. The quality score ranges 0 to 56 from low quality to high quality. 55.50 is definitely a good score, while 13.80 might be not. Nevertheless, sequences with low quality may still be true and map to the genome nicely.
2) .x means vector sequence found and removed. Does this mean that some of the
binary vector used for transformation goes along with the T DNA and it comes
after sequencing with LB primer? let's say, in order: LB T-DNA, vector
sequence, genomic sequence?
A: The vector sequence is
either the T-DNA sequence or the adoptor sequence, or both. The order should be:
3) L300-3' This means that the insertion is located in the last 300 bases
of the gene, at its 3' end. Could be that is actually in the last exon?
or in the 3'UTR? And then to double check i need to use PCR?
LB -- T-DNA -- genomic sequence -- adoptor sequence.
20. Q: How do I acknowledge Salk sequence-indexed
insertion lines in talks or publications?
A:
21. Q: Are there any other ways to access the information
in addition to SIGnAL T-DNAExpress page?
A: A: (Siebert,P.D., Chenchik,A., Kellogg,D.E., Kukyanova, K.A. and
Lukyano, S.A (1995) An improved PCR method for walking in uncloned
genomic DNA NAR 23: 1087-1088
A: A: A: A: A: A: A: A: A: A: A:
A: A: A: A: Nevertheless, you could retrieve the FST from our site. If the FST was not trimmed, you could blast align them on the genome and then plus the start mapping bps to estimate the actual insertion site. Due to lack of information if the FST is trimmed or not, we did not provide information of the estimate site.
In your case, the region 504-737 bps of the FSTs were mapped on the genome, that is, its actual insertion site is at least 500 bps away in the downstream of the genome, if the FSTs was not trimmed and the vector sequence was removed. The insertion site is estimated as 23464585+504 = 23465089, which falls in the 6th intron of the gene
( W/23463915-23464130,[1] 23464212-23464282,[2] 23464381-23464395,[3] 23464512-23464623,[4] 23464712-23464811,[5]
23464897-23464959,[6] 23465184-23465294,23465373-23465557,......).
A: http://www.arabidopsis.org/servlets/SeedSearcher?action=detail&stock_number=SALK_055016C
the see means homozygous. .but you can ask for segregating populations too...
A: The c/ group has no difference with w/ group with respect to their LB primers, as well as its LP or RP. The iSect Primer tool has already adjusted to recognize the insertion dirction, thus LB + RP will be always the combination for the insertion PCR, while LP+RP for the Wile Type.
There is a description in the web page:
"By using the three primers (LBb1+LP+RP) for SALK lines, users for
WT (Wild Type - no insertion) should get a product of about 900-1100
bps ( from LP to RP ), for HM (Homozygous lines - insertions in both
chromosomes) will get a band of 410+N bps ( from RP to insertion
site 300+N bases, plus 110 bases from LBb1 to the left border of the
vector), and for HZ (Heterozygous lines - one of the pair
chromosomes with insertion) will get both bands."
However, I got 2 bands for the homozygous mutant instead of one when
I using the three primers together. There is always a strange band
with a size nearly 800 bp appeared in the mutant sample. I wonder if
the primer suggested here have some problems (for the LBb1 I am
using ) or it is just normal to got three bands with different size
in the reaction.
A: In addition to switching to LBb1.3, I would suggest that you don't
combine
all three primers in a single reaction. This saves an extra
reaction but
can also result in artifact bands. Instead, you might want to run an
LeftPrimer-RightPrimer pair , and a RightPrimer-LBb1.3 pair as two
seperate reactions on the DNA sample that you are genotyping.
A: 42. Q: We have recently used your lab's adapter ligation-mediated PCR
technique to explore several unusual T-DNA insertions, and I had a
few questions about how the protocol would need to be modified in
order to investigate T-DNA right-border junctions. Have you ever
used this technique in this manner (using right-border T-DNA
primers)? Can you recommend any primers which were particularly
successful? If you have not, can you recommend any changes to the
protocol that would be necessary for this to work?
A:
The primers we used for the right border rescues were:
JMRB2 "TGATAGTGACCTTAGGCGACTTTTGAACGC" (this is the primer for the
1st PCR)
JMRB1 "GCTCATGATCAGATTGTCGTTTCCCGCCTT" (this is the primer for the
2nd PCR, nested to JMRB2)
We did not get very good results for the rescue of the right
borders. In our experience we used to get tones of nice PCR
products, but MOST of them contain no Arabidopsis DNA (they were
just vector sequences).
After few hundred sequences and having tried few different
restriction enzymes, we decided that it was not worth it for our
purposes.
This does not mean you should not try. I think you can increase your
chances of getting the RB insertion sites by using not one but
several restriction enzymes (one at the time).
43. Q: My question in a nutshell is: Where exactly is the left and right border of the PROK 2 Vector?
A: Paul Shin, the sequencing coordinator, and Chris Kim, the lab manager, had made great efforts on finding the exact left border and the right border were. Moreover, we also re-sequenced the vector. The pBIN-pROK2.txt-new¿ was their last result. You could find where were the left border and the right border of the insertion in this file. Nevertheless, the txt-new was our re-sequencing result and it might be different from other's vector sequences published.
The 13 QA was to answer left border primer question and no left border information was there.
I have just revised the answer and extend the sequence to the left border to avoid confusion.
Even though the exact left borders for majority of the T-DNA insertions were proved the same as txt-new, some, significant number, did not exactly agree to txt-new. The right border was more complicated story. Many could be found for their border, but more not. There were also many insertions with double copies of T-DNA together. At first, we were also trying to locate the insert sites by using the right border primers, but the results were much worse than using the left border primer due to these reasons.
We could design primers easily if we could locate the exact insertion site and the insertion sizes were constant. Our results from using NG are much closer to the exact sites (http://neomorph.salk.edu/T-DNA_Seq.php).
44. Q: I have questions about SALKSeq T-DNA lines. What is the relationship between them and SALK lines? Does it mean that SALK_130432 line (available at NASC) is exactly the same than the SALKseq_130432.1 line later described?
A:
The 'SALKseq' records are not a separate set of lines, they are just records of identified insertions in the existing SALK lines. The SALKseq project is a re-sequencing of the existing lines using next-gen sequencing. The six digit number after SALKseq_ , (e.g. 130432) corresponds to the number of an existing line (e.g. SALK_130432). The trailing decimal number is the insertion number in that stock, so if multiple insertions are identified in a stock they will be named SALKseq_XXXXXX.0, SALKseq_XXXXXX.1, etc. For any given SALK line, it will have one or more insertion identified by sanger sequencing (e.g. SALK_130432.38.30.X) and possibly some SALKseq insertions.
It's worth noting that a SALKseq record may correspond to the same physical insertion as a regular SALK record. For example, SALK_130432.38.30.x and SALKseq_130432.0 both refer to the same physical insertion in AT5g65560. In this case, the SALKseq record serves as additional validation of this insertion.
On the polymorphism page that you included in your email, there is a section labelled "Germplasm" which contains a link to the seed line(s) that contain the SALKseq polymorphism. Since these are the same familiar SALK lines they should be available at NASC, and you can still use the LBb1.3 primer.
45. Q: I understood that the name of SALKseq should be related to name of original SALK line and if you'd got more than one sequence so it suggested more than one T-DNA insertion site in particular original SALK line. Am I correct?
A: Yes, you are right. If you'd got more than one sequence so it suggested more than one T-DNA insertion site in particular original SALK line. 46. Q: I found information in publication Su and Krysan 2016 that they used primer p745 for genotyping of SALK lines. Please could you confirm or reject this info? I thought the primer p745 is for Wisc lines only. Could it be used for genotyping of SALK or SAIL lines?
(Su and Krysan 2016. A double-mutant collection targeting MAP kinase related genes in Arabidopsis for studying genetic interactions. Plant J. 2016 Aug 4. doi: 10.1111/tpj.13292.)
A:
To standardize our genotyping pipeline, we mainly used T-DNA primer LBb1.3. However, our portal also allows researchers to design their own T-DNA primers as stated in Q&A #3 and using our iSec Tools page :
http://signal.salk.edu/isectprimers.html
47. Q: I was wondering if you could let me know the best T-DNA specific primer to use to genotype the WiscDsLox lines. I found a T-DNA specific primer for the WiscDsLoxHs lines (L4; TGATCCATGTAGATTTCCCGGACATGAAG), but I assume that I would need a different primer for the WiscDsLox lines. I found one reference to primer P745 (AACGTCCGCAATGTGTTATTAAGTTGTC). Would that be the proper primer for the WiscDsLox lines? Thanks for your help.
A:
The border primers that we were using can be found in the section "3.3 Designing genotyping primers with the iSect tools" of our user's guide https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5215775/.
1) we submit all sequences to Genbank (GSS division
http://www.ncbi.nlm.nih.gov:80/dbGSS/). (the identified "TDNA" can be
use to locate all entries in a text search).
2) we provide all sequences to TAIR - they provide a blast service
with links to the available stocks.
http://www.arabidopsis.org/abrc/tdna_ecker.html
22. Q: What method do you use to recover the plant flanking T-DNA
sequences in the Salk insertion lines?
23. Q: Where can I get the pROK vector?
24. Q: Is the insertion point in the tdnaexpress page the exact insertion site?
25. Q: We have been using the SALK lines to obtain WRKY knockout lines. In the
course of our analyses we often have encountered that the LBb1 primer
alone gives a clear 450 bp PCR product even under relatively srtringent
PCR conditions (60°). Naturally, we assumed that we have some
contamination problem. However, yesterday a completely independent lab
at our institiute working with another gene family and with their own
primers told us they observe exactly the same (I saw their gels and I am
certain it is the same product). Have you seen this or has othrs
reported this to you?
26. Q: First, there are two SALK_009273:009273.19.95.x and 009273.56.00.x
They are both at the same position on At5g65430, but according to T-DNA
Express, the T-DNA insertions are in opposite directions. Does it mean that
I will have to run PCRs in both directions?
27. Q: I received T-DNA express SALK-006294.
I don't know what high e-value mean. Please, let me know how can I understand " Caution: high e-value"?
For exmaple:
SALK_116614.18.85.x [seq]
CHRO chr1
POSN 8392
DIRE C/8293-8392
VALU 7e-07 Caution: high e-value.
HITS At1g01020
COD2 T25K16.2
TITL unknown protein; protein id: At1g01020.1 [Arabidopsis thaliana]
LOCA Intron
COOR C/7729-7835,7942-7987,8236-8325,8417-8464,8571-8666
28. Q: while looking for T-DNA insertions for CODE At1g62830,
I obtained names of different hits with the same clone number. For example:
SALK_048276.55.75.x, SALK_048276.23.95.x and SALK_048276.40.90.x.
29. Q: I have used the software in the web page to find suitable RP and LP and it gave me this result:
SALK_029701 PRODUCT_SIZE: 977:
LP: gcagctgcatcaggttcgtct LENGTH: 21 TM: ....
RP: ccccttttcttcgttcgcatc LENGTH: 21 TM: ....
When I take the my whole gene sequence and I can not find the
regions where the primer where designed by the software (at least I
should one of them). But when I paste the result of the primer
design in the T-express search it give me the right thing.
30. Q: Is it correct that MAX N should be 500 instead of 300 for design of primers to work with LBa1? I would think that MAX N should be 100 in
these case to preserve the size of the band for HM, which will be 300 + 310 +N = approx.700. 310 in this equation is a distance between LBa1 and left border (110+200) if LBa1 locates 200bp further than LBb1. Is it right?
If HM and using LBa1, the new product size is estimated:
200+110+(N-200)+300 = 410+N ~= 910
LBa1 LBb1 LB FS RP
| | | | |
--------------------------------------------->
| 200 | 110 | N-200 | 300 |
If no insertion, the genomic product size is estimated:
300+N+300 = 300+200+(N-200)+300 = 600+N ~= 1100
LP LB FS RP
| | | | |
------------------------------------------------->
| 300 | 200 | N-200 | 300 |
| N |
LP -- left primer
RP -- right primer
FS -- flanking sequence start point
LB -- the insertion site (Left Border)
LBa1, LBb1 -- the LB primer
31. Q: When I was looking for T-DNA insertions for CODE At3g28290, I obtained
names of different hits with the same clone number, SALK_070418.25.45.x, SALK_070418.35.40.x.
But when I looking for At5g41800 ,I also found SALK_070418.20.10.x. Now I have the seeds of SALK_070418,
but how I know which gene the seeds should belong to? Use PCR or other methods?
For the line SALK_070418, we sequenced it three times because its
sequencing scores were lower than 30 at the first and second times. Obviously this line might have two
insertion sites, one in chr3, the other in chr5. You could use http://signal.salk.edu/tdnaprimers.html to generate
primers. The page would return 2 pairs of primers (two sequences share one pairs). You could use them
with LBa1 or LBb1 to set up two PCR reactions. By combining the results from two reactions, you could
identify if a line is with two insertions or one insertion, as well as is HM, HZ or WT.
32. Q: Are SAIL lines (formerly GARLIC) now deposited into your databse? If so, what is the binary vector sequence and the prospcetive LB primers used to
sequence? If not, where would I obtain information in that regard?
33. Q: I have some SALK T-DNA insertion lines that I am trying to genotype
using PCR. I have been using the isect toolbox website to design
primers. I am a little confused about the distance from the primer
LBb1 to the insertion site. On the isect website it describes the
distance as 110 bp. However, when I look at the pBIN-pROK2 map from
the website it looks like LBb1 locates 216 bp from the left border. I
was just curious where the 110 bp number came from?
> pBIN19
Score = 58.2 bits (23), Expect = 5e-12
Query: 1 tggttcacgtagtgggccatcg 22 LBa1
||||||||||||||||||||||
Sbjct: 6481 tggttcacgtagtgggccatcg 6458
Score = 55.7 bits (22), Expect = 3e-11
Query: 1 tcaaacaggattttcgcctgct 22 LB6313
||||||||||||||||||||||
Sbjct: 6313 tcaaacaggattttcgcctgct 6292
Score = 55.7 bits (22), Expect = 3e-11
Query: 1 gcgtggaccgcttgctgcaact 22 LBb1
||||||||||||||||||||||
Sbjct: 6280 gcgtggaccgcttgctgcaact 62
34. Q: I have used the TDNA verification primer design tool in the past and it has worked great. However it won't design primers for: SAIL_748_E04 and SAIL_748_B04.
Do you know why or have any suggestions?
SAIL_748_E04 Insertion chr3 4467442 No Primer Found. Please edit parameters and try again.
PRIMER PICKING RESULTS FOR SAIL_748_E04
Using 0-based sequence positions
NO PRIMERS FOUND
SEQUENCE SIZE: 1101
INCLUDED REGION SIZE: 1101
EXCLUDED REGIONS (start, len)*: 101,899
0 GATGTTAATTTTTTTGATATATTAAACTTTTTATAAATTTAAATATTAATAGTGTTTTAT
60 GAATTTTAAATATATAAATAATAATAATTAAAAAATTAATAttttttgccttttaggtaa
XXXXXXXXXXXXXXXXXXX
120 tgatataataattctaaaagcaagaatttagaagatttaaatctttaattaaatattttc
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
180 atgcctgaataattaatgtcagtagcatgacattgtaaataagttcaaatacatgatttg
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
240 tttatttaaacaaaaaacgtttagtaagaacaatatttatttaaacaaaaaactttgttt
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
300 taaaatattgttaacaaaatacttttatgctaattttaaaggattgatgtgcagtttgtt
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
360 tggattaaatttgattgataatgtgattgacaatttaaatTAAGAATTTAATGTAAAATT
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
420 AATTATTTTGAAAAAAAAATTTAATGCTAATGGCATTGGGTTGTAAATAAGTTCAACTCT
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
480 AAGATTTATTTGTTAAAATGGCTGCAAAAATGTATATGTAGATTGGCAACACCGGCCTTG
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
540 AGATCAAACGACTTGGTGCGAGCCTCGACATAACCTCTAGCAAATCGAAAGATTCCACTA
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
600 CCTCCGACCACCGGCATTTCCCTAACCTTCGACATCACCACGTTCCGCCCAAGAATCGTT
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
660 ATCGTACTCCCATTGTACTTCCCCGTAGTGAAAGCAAAGTTcatcaccatcaagaagcct
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
720 atctctccttgggccgctgctacgtacatcccttgggcttggcccaccacagtcgcattc
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
780 cttggcacatcgaatgttagtggatcgtccatcatggtgatcgatccggataaggaagag
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
840 ttcaagaccggctgttggatcatcacggagctagggtttctaccgtttacagagttgtgc
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
900 caatagactcggagatgagtgagtttctcctttttgtaagggccaaggtgtttcccgttc
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
960 atggttcttcccaattcatctccgacggaagcaacggcatAGAAAAGGAGGATTTGTGCG
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX
1020 GCGAAGATAAGGATGAGGTTAGTCATGTTGTGGTTTTTGTGTCTCTAGATGAAGGTTTGT
1080 GCTATAAATAAACGAGTGTTT
KEYS (in order of precedence):
XXXXXX excluded region
35. Q: If could I share my exprimental analysis data with other scientists for specific lines?
36. Q: If all SALK Homozygous lines contain only one T-DNA insertion?
37. Q: I was bugging one of our user to give us the accession id of the mutant allele they had been using. They said it is SAIL_390_C01. However, in the process of retrieving the info, the found out that this line is reported on the SiGnal site to be located either in intron 2 or exon 4. The information they had published (that had been obtained from Syngenta through a Blast) said that the insertion was in intron 6. Moreover, the researcher had sequenced the insertion and found that it was indeed in intron 6. Do you know why it appears at a different location in the SiGnal DB?
38. Q: Dear SIGnal team, I need three T-DNA lines, but I can't find these three lines anywhere:
Is it possible to get them over you directly?
SALK_055016.53.30.x
SALK_099132.30.45.x
SALK_031802.55.25.x
SALK_055016
SALK_099132
SALK_031802
39. Q: I am trying to design primers for a SAIL line. In regards to the
insert primer, you provide 3 primer sequences (LB1-3). How do I determine which one to use for my line?
There is no info on which "C/" group a particular line belongs to. If these were the primers used for
Syngenta's TAIL PCR, should we use LB3 for any SAIL line?
40. Q: I am using the primers suggested in the web page:
http://signal.salk.edu/tdnaprimers.2.html and I encountered a problem.
41. Q: I need to ask some questions about the SALK insertion lines.
Is the insertion of the t-dna in only ONE position within the affected
gene or can there be multiple insertions within the affected gene?
2. Each SALK line corresponds to a specific gene, is this correct?
I know that some genes have more than one SALK line associated with them.
3. Is it correct to assume that insertions can be present in other
parts of the genome of a given SALK line besides the affected gene?
If I go to this page of you website: http://signal.salk.edu/pBIN-pROK2.txt-new
it shows the left border at 6042
5941 GTTTCAAACCCGGCAGCTTAGTTGCCGTTCTTCCGAATAGCATCGGTAACATGAGCAAAG
6001 TCTGCCGCCTTACAACGGCTCTCCCGCTGACGCCGTCCCGGACTGATGGGCTGCCTGTAT
|<-- Left border
6061 CGAGTGGTGATTTTGTGCCGAGCTGCCGGTCGGGGAGCTGTTGGCTGGCTGGTGGCAGGA
On your FAQ page, the left border appears to starts at 6121.
In the 13. Q: What is the DNA sequence around
the left border of pROK2 and where do the PCR isolation and DNA sequencing
primers anneal?
A: The sequence of the left border of pROK2 and the position of the PCR amplification and sequencing
primers:
6121 5'-TATATTGTGGTGTAAACAAATTGACGCTTAGACAACTTAATAACACATTGCGGACGTTTT
6181 TAATGTACTGGGGTGGTTTTTCTTTTCACCAGTGAGACGGGCAACAGCTGATTGCCCTTC
6241 ACCGCCTGGCCCTGAGAGAGTTGCAGCAAGCGGTCCACGCTGGTTTGCCCCAGCAGGCGA
3'<<<<<<<<<<<<<<<<<<<<<< 5'
When I enter the PROK2 vector sequence from your website into Snap Gene, an annotation (LB T-DNA repeat) appears at 6113.
These aren't huge differences, but I'm really curious, where exactly is the left border, at 6042, 6121, or 6113?
And why is there an apparent discrepancy?
I am a bit confused and would appreciate so much a bit of help/further info
Plus, most of people is not familiarized with this SALK Seq lines yet..is the LbB1.3 primer useful to genotype these lines too? Or you recommend another version?
Your FAQ say 'The six digit number after SALKseq_ , (e.g. 130432) corresponds to the number of an existing line (e.g. SALK_130432). The trailing decimal number is the insertion number in that stock, so if multiple insertions are identified in a stock they will be named SALKseq_XXXXXX.0, SALKseq_XXXXXX.1, etc.' however many of your SALKseq names cannot match existing SALK line names because they are shorter. Example - SALKseq_70256.2 has 5 digits for identification but SALK lines have 6 digits in name so - what SALK line match this SALKseq with 5 digits only? And many more SALKseq records have 5 digits only.
Yes, partially. 'The six digit number after SALKseq_ , corresponds to the number of an existing line'. However, not all SALKseq lines have SALK numbers. Since we used the single path sequencing strategy to retrieve FSTs of SALK lines before NG, we could not get high quality FSTs for many lines with disturb due to multiple sites or other reasons. Salk lines with FSTs were assigned SALK numbers and then submitted to NCBI and ABRC. Nevertheless, those Salk lines without FSTs had no SALK numbers and were not submitted to ABRC. By using the NG sequencing, we could get sequences from new sites and new lines. We got 11,022 sites from 6,270 lines which we could not get any before. These lines had no SALK numbers and were treated as new lines or original lines. They were assigned SALKseq_ with 5 digits or less, and submitted to ABRC.
Primer p745 used by the authors maps to pBIN-pROK2 plasmid used to generate the Salk T-DNA collection.
p-745 5'-AACGTCCGCAATGTGTTATTAAGTTGTC-3'
>pBIN-pROK2
6121 TATATTGTGGTGTAAACAAATTGACGCTTAGACAACTTAATAACACATTGCGGACGTTTT
<<<<<<<<<<<<<<<<<<<<<<<<<<<< p-745
6181 TAATGTACTGGGGTGGTTTTTCTTTTCACCAGTGAGACGGGCAACAGCTGATTGCCCTTC
6241 ACCGCCTGGCCCTGAGAGAGTTGCAGCAAGCGGTCCACGCTGGTTTGCCCCAGCAGGCGA
<<<<<<<<<<<<<<<<<<<<<< LBb1
6301 AAATCCTGTTTGATGGTGGTTCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGC
<<<<<<<<<<<<<<<<<<< LBb1.3
6361 CCGAGATAGGGTTGAGTGTTGTTCCAGTTTGGAACAAGAGTCCACTATTAAAGAACGTGG
6421 ACTCCAACGTCAAAGGGCGAAAAACCGTCTATCAGGGCGATGGCCCACTACGTGAACCAT
<<<<<<<<<<<<<<<<<<<<<< LBa1
In Table 1 we provide a list of collection-specific T-DNA left border primers that we have regularly used in-house.
Table 1 : T-DNA specific primers for each of the insertion mutant collections
Collection PrimerID Primer sequence
SALK LB-1.3 ATTTTGCCGATTTCGGAAC
SAIL LB-1 TAGCATCTGAATTTCATAACCAATCTCGATACAC
WiscDs LB TCCTCGAGTTTCTCCATAATAATGT
WiscDsLoxHs L4 TGATCCATGTAGATTTCCCGGACATGAAG
GABI-Kat o8409 ATATTGACCATCATACTCATTGC
