Frequently Ask Questions:
1) How do I search for my favorite gene?
2) How can I tell if a gene is a possible CREB target by looking
at CRE prediction?
3) How do I know if a promoter is bound by CREB in the ChIP-on-chip
data?
4) How can I determine if a gene is induced by cAMP in a specific
tissue by looking at microarray data?
5) Who should I contact if I have additional questions or comments?
6) How to cite the database?
7) How do I use the LinkOut feature to go directly from NCBI gene
to the CREB target gene database?
1) How do I search for my favorite gene?
You can use the gene symbols, gene names, Genbank accession numbers or locuslink
numbers to search the database. We recommend using the official gene symbols
or gene IDs (locuslink numbers) to perform the search. If you are not sure about
the official gene symbol or gene ID of your gene, please go to NCBI
Entrez Gene and do a search there.
The default search returns the CRE prediction data, with links to all other
databases (through locuslink numbers)
2) How can I tell if a gene is a possible CREB target by looking
at CRE prediction?
If it's a "CRE_TATA" or "CRE_NoTATA" gene, there is a good
chance it is a CREB target. You are also encouraged to check the location of
the CREs, especially if the CRE is not conserved; generally those that are located
close to transcription start site are more likely to be real sites. A few CREs
located near each other are also a good sign for CREB binding. The presence of
downstream TATA box is a good indication of cAMP inducibility.
3) How do I know if a promoter is bound by CREB in the Chip-on-chip
data?
For each Chip-on-chip experiment, p-values and binding ratios are given. We
use p-value <=0.001 and binding ratio>=2 as cut off for CREB binding.
Please note the sensitivity of Chip-on-chip using this cutoff is about 50%.
If you see a gene with binding ratio somewhere between 1.5 and 2, there is a
good chance that it may be a CREB target and manual ChIP might be needed to
verify it.
4) How can I determine if a gene is induced by cAMP in a specific
tissue by looking at microarray data?
Our microarray data is processed by dChip. The fold change (FC) value is positive
for increased expression and negative for decreased expression. There is also
a lower bound of fold change (LFC), which is a conservative estimation of the
fold change with 90% confidence. We recommend using the lower bound of fold
change to determine changes in expression level; if cAMP induced a fold change
with LFC>=1.5, it is a good induction, and you can use looser criterion as
LFC>=1. For HEK293T and MIN6 cells, we also have ACREB experiments. If the
dominat negative ACREB caused a reduction of LFC<=-1, it is a good indication
that the effect of FSK is CREB dependent.
5) Who should I contact if I have additional questions or comments?
Please drop a message to Xinmin Zhang.
If you found the database useful in your work, please cite our PNAS paper:
Genome-wide
analysis of cAMP-response element binding protein occupancy, phosphorylation,
and target gene activation in human tissues
Xinmin Zhang, Duncan T. Odom, Seung-Hoi Koo, Michael D. Conkright, Gianluca
Canettieri , Jennifer Best, Huaming Chen, Richard Jenner, Elizabeth Herbolsheimer,
Elizabeth Jacobsen, Shilpa Kadam, Joseph R. Ecker, Beverly Emerson, John B.
Hogenesch, Terry Unterman, Richard A. Young, and Marc Montminy
Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4459-64. Epub 2005 Mar 7.
If you used the rat CREB binding data (SACO) or PC12 gene expression data,
please cite:
Defining
the CREB regulon: a genome-wide analysis of transcription factor regulatory
regions. Cell. 2004 Dec 29;119(7):1041-54.
Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney
S, Dunn JJ, Mandel G, Goodman RH.
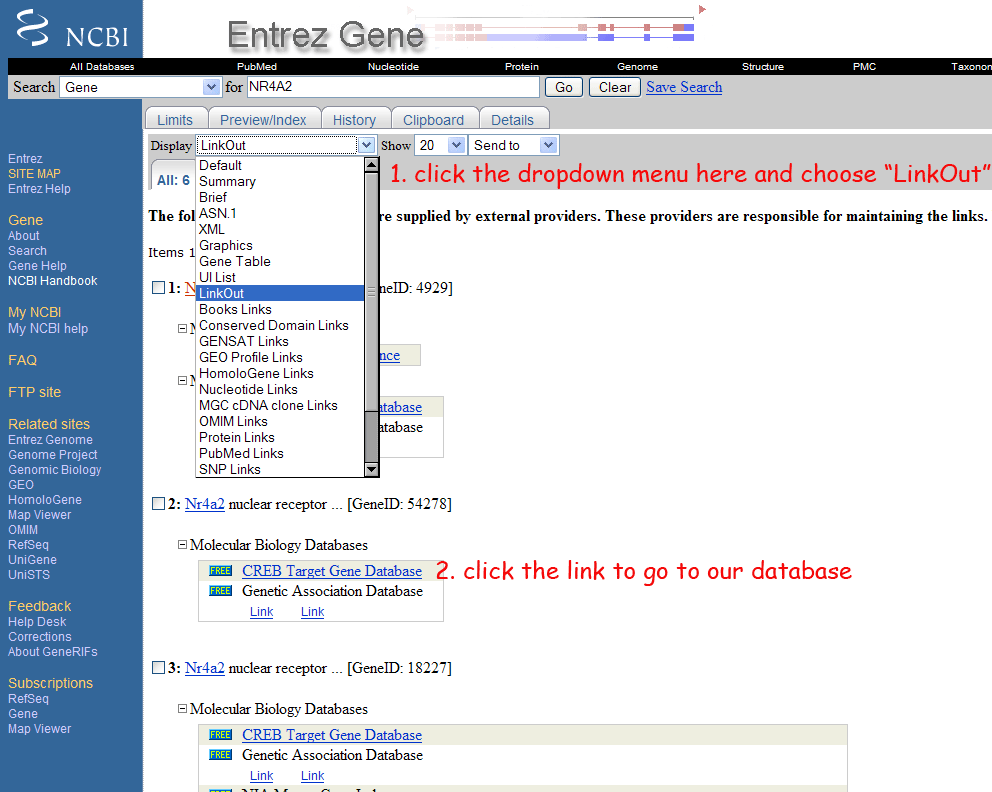
7) How do I use the LinkOut feature to go directly from NCBI gene to the CREB target gene database?
Once you've found your favorite genes from NCBI Entrez Gene, choose LinkOut in the display options. If that gene is in our database, a link will be shown and you can click that to go to our database (see the image below).
A complete list of genes that are linked from NCBI to our database can be found here.
