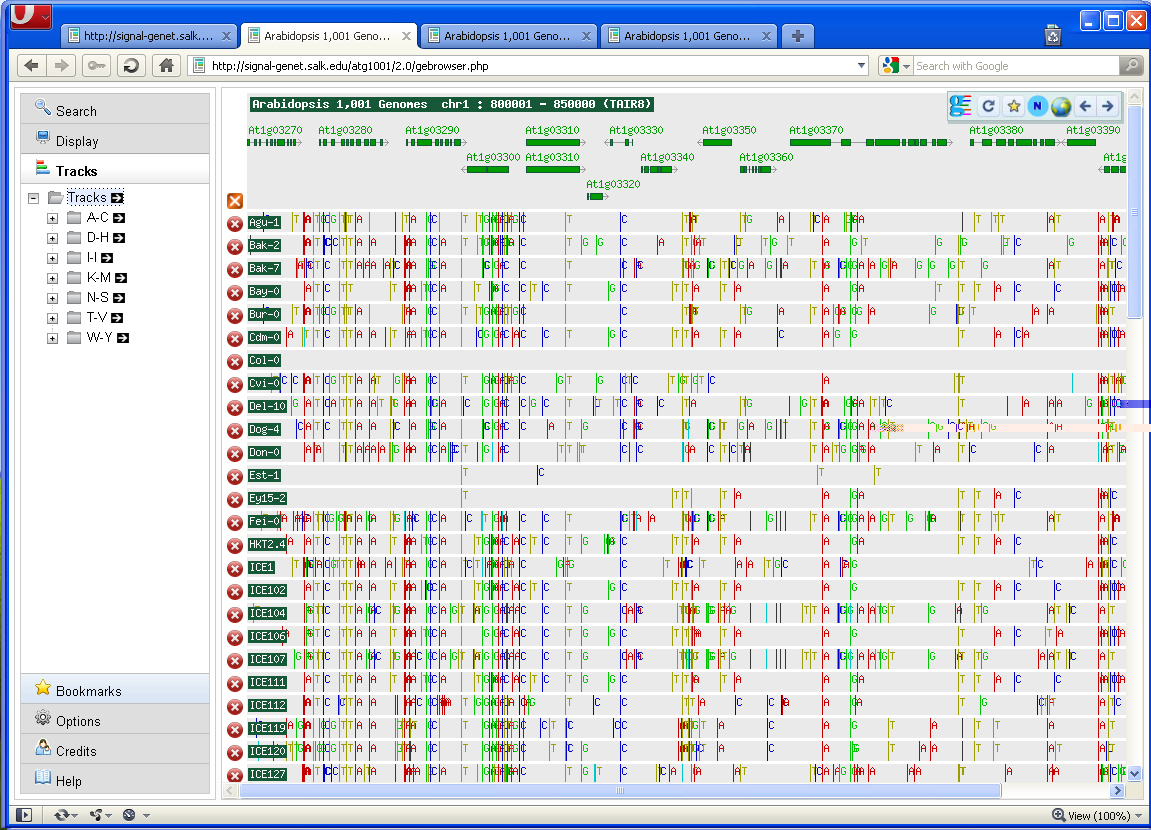
Screen Shot
Search
Locus: Type in AGI# (e.g. At1g01040) and click the refresh arrow  . If
multiple splice forms are present for a locus then a specific form
will need to be selected (e.g. At1g01060.1, At1g01060.2, etc.)
. If
multiple splice forms are present for a locus then a specific form
will need to be selected (e.g. At1g01060.1, At1g01060.2, etc.)
Display
Chr: Select a chromosome number.
Posn: Select a position for the chromosome selected above.
Zoom: The zoom feature allows for specification of window size.
Length: This feature will adjust the browser window size (if the length is increased beyond the size of the browser then a scroll bar will appear on the bottom of the browser page).
Code: The code has two options (Nucleotide and Amino acid). If [Nucleotide] is selected then nucleotide substitutions will be displayed in the browser. Same for [Amino acid] Be sure to refresh the screen after switching the defaults by clicking on the green refresh arrow.
View: This feature allows the user to toggle between a [Graphic] or a
[Text] view of the [Nucleotide] or the [Amino acid] code selected
above. Be sure to refresh the screen after switching the defaults by
clicking on the green refresh arrow  .
.
Tracks
Accessions have been binned alphabetically. Accessions can be loaded
into the browser by clicking on them. All accessions from a
particular bin (e.g. A-C) can be loaded into the browser by clicking
on the black arrow  immediately to the right of the bin name.
immediately to the right of the bin name.
Accessions can be removed individually by clicking on the closing button  within a
circle next to the accession name. In addition, clicking on the larger closing button
within a
circle next to the accession name. In addition, clicking on the larger closing button
 located in the top left of the browse will remove
all accessions.
located in the top left of the browse will remove
all accessions.
Bookmarks
The bookmark feature allows the user to save selected loci under
investigation. Type in a label (e. g. AGI#) and click on the add bookmarks button  to store the
locus. A bookmark can be removed by selecting on the small removal button
to store the
locus. A bookmark can be removed by selecting on the small removal button  option or All on the larger button
option or All on the larger button  option.
option.
Options
Bookmarks: Selecting this option will enable any AGI# selected in the browser to be automatically stored under the bookmarks tab.
SAAP: (Single Amino Acid Polymorphism) - There are two options for this feature (All and CDS only). Selecting [All] will display all amino acid changes regardless of their location within a gene. Selecting [CDS only] will display amino acid polymorphisms within the CDS. Nucleotide polymorphisms that result in a synonymous amino acid will be displayed with a green flag; non-synonymous changes will be displayed with a red flag. [Important Note] When using the [CDS only] option only one specific locus can be interrogated at a time. Scrolling to the next locus or the next window will restore the default [All] option and ignore the [CDS only] choice.
Locus display: Enabling this option will display the full and exact length of a gene if the user clicks on a specific AGI# within the browser.
Label Alignment: Changing the alignment of the strain accession label.
View: Checking this [Toolbar] option will launch the toolbar pane on the upper right of the window to allow quick operations below.
![]() ==> equavelent to
==> equavelent to 
![]() ==> bookmarks
==> bookmarks  .
.
![]() or
or ![]() ==> set [Display]->[Code] to [Nucleotide] or [Amino Acid] +
==> set [Display]->[Code] to [Nucleotide] or [Amino Acid] + 
![]() or
or ![]() ==> set [Display]->[View] to [Graphic] or [Text] +
==> set [Display]->[View] to [Graphic] or [Text] + 
![]() ==> display previous or next screen of chromosome walking.
==> display previous or next screen of chromosome walking.
Legend
Nucleotide
A SNPs -- Red line
C SNPs -- Blue line
G SNPs -- Green line
T SNPs -- Yellow line
1 bp deletions -- Black line
1-3 bp insertions -- Purple Triangle (w /), [AG], or [f+1](AA) (+1 frame shift 1 bp, // frame shift 2 bps.)
Unsequenced regions -- .. (dot) or grey area
Depending on the zoom some SNPs may appear in Teal which actually represent mulitple SNPs in close proximity to one another. These can be resolved to the actual SNP by zooming in or switching the Code to "Text".
Amino Acide
Synonymous amino acid -- Green line
Non-synonymous amino acid -- Red line
All standard amino acide codes are used. In addition, a Z - indicates an unknown amino acid, possibly a deletion and an X - indicates an amino acid has become a stop codon.